Elena Canorea
Communications Lead
During the last few years, a research effort has been made to automate the analysis of medical images used in the diagnostic-therapeutic cycle.
In modern medicine, making a diagnosis using images is invaluable. Computer tomography (CT) imaging and other modalities provide a non-invasive and effective means of delineating the subject’s anatomy.
Image segmentation has become a key process for the delineation of certain anatomical structures and other regions to assist and aid physicians in surgery, biopsies, and other clinical tests.
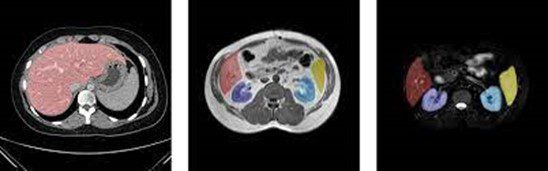
Example of organ segmentation
Over time, Artificial Intelligence techniques, and in particular the use of Deep Learning, have allowed the field of computer vision to advance rapidly in recent years. In this article, we show how we can use semantic segmentation techniques for organ detection in medical images.
Currently, convolutional neural networks have been used to solve different tasks within the domain of Computer Vision. The resolution of each of these tasks provides us with a different degree of understanding of the images.
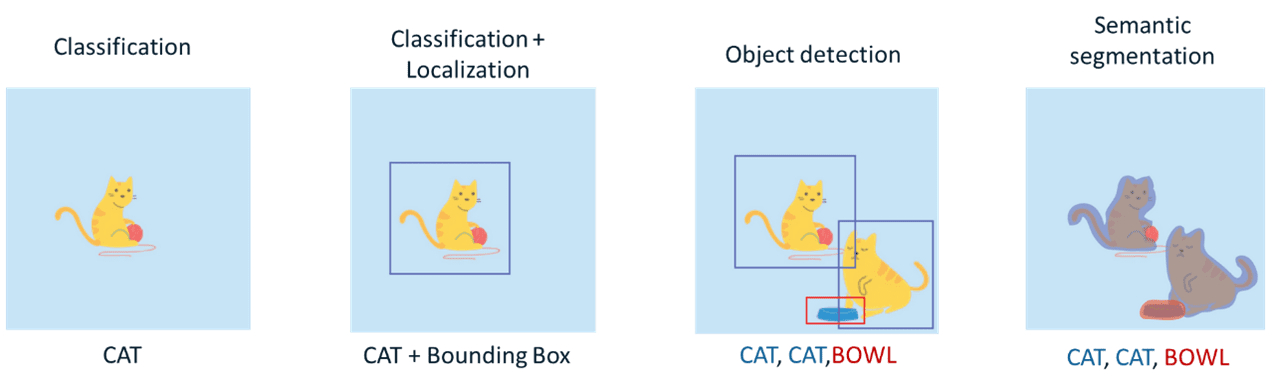
Types of Computer Vision tasks
As previously mentioned, in this case, we will focus on one of the most complex tasks in the field of computer vision: Semantic Segmentation. The goal of this task (within the field of images) is to label each pixel of an image with the corresponding class of what it is representing. Because we are performing prediction for each pixel in the image, this task is commonly referred to as dense prediction.
The performance of our model will largely depend on the quantity and quality of the data set. For this reason, the selection of the dataset is one of the most important processes within the workflow of Machine Learning projects.
As mentioned at the beginning of the article, the goal of our model is to detect organs (at the semantic segmentation level) from medical images of the abdomen.
Due to the sensitivity of this medical information, an anonymized dataset (CHAOS: Combined Healthy Abdominal Organ Segmentation) Dataset) has been used specifically for research topics within this area. This dataset is composed of several sources. In this case, we have focused on using a subset of data for the detection of the following organs: spleen, kidneys (left and right), and liver.
Throughout this section, we will discuss the most important aspects that have been conducted during our organ segmentation training process.
As our dataset consists of DICOM images, before launching the training process, it is necessary to process all our data and convert these images into an array of pixels. Once the array extraction is done, several transformations will be performed: affinity and image size reduction. Finally, the images are normalized in order to obtain a better performance during the training phase.
U-Net is a convolutional neural network architecture that was developed for biomedical image segmentation, therefore, we will rely on its base architecture for our model definitions. On this occasion, we have used some variants of the original topology to perform our training process: U-Net with (Resnet18) and U-Net++.
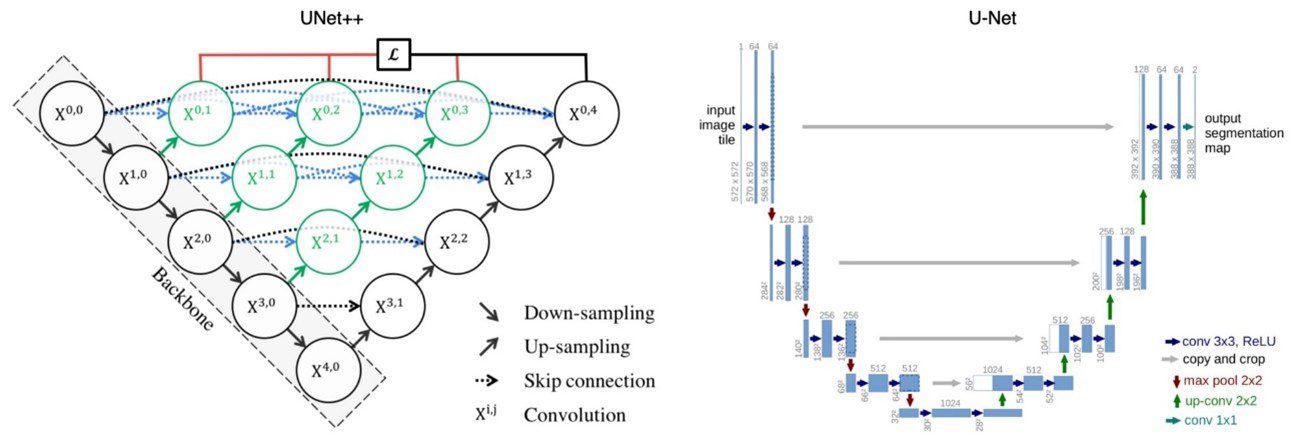
Topology of the models used
The loss function in the field of neural networks is a function that evaluates the deviation between the predictions made by the network and the real values of the observations used during the training process. The lower the result of this function, the more efficient the neural network is. The objective of training a neural network is to try to minimize this function as much as possible in order to minimize the deviation between the predicted value and the actual value.
One of the most commonly used loss functions in the case of image segmentation is the “dice” coefficient. This coefficient is used to determine the degree of overlap between the predicted mask and the labeled mask.
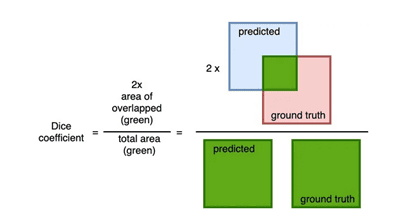
Calculation of the “dice” coefficient
The Dice coefficient has a range of values between [0,1] where 1 indicates a total overlap. Therefore, if we want to minimize this function as much as possible, the value of (1- Dice coefficient) can be used as a loss function.
In our case, the final loss function used for training is a combination of the Weighted Binary Cross-Entropy loss function and the Dice loss function.
In this section, we will discuss the results obtained during the training process.
The hyperparameters used in the training were the following:
As we can see in the following image, the Unet-Resnet model converges faster than the Unet++ model (this is due to the use of a model with pre-trained weights in the “encoder” layers and a smaller number of parameters).
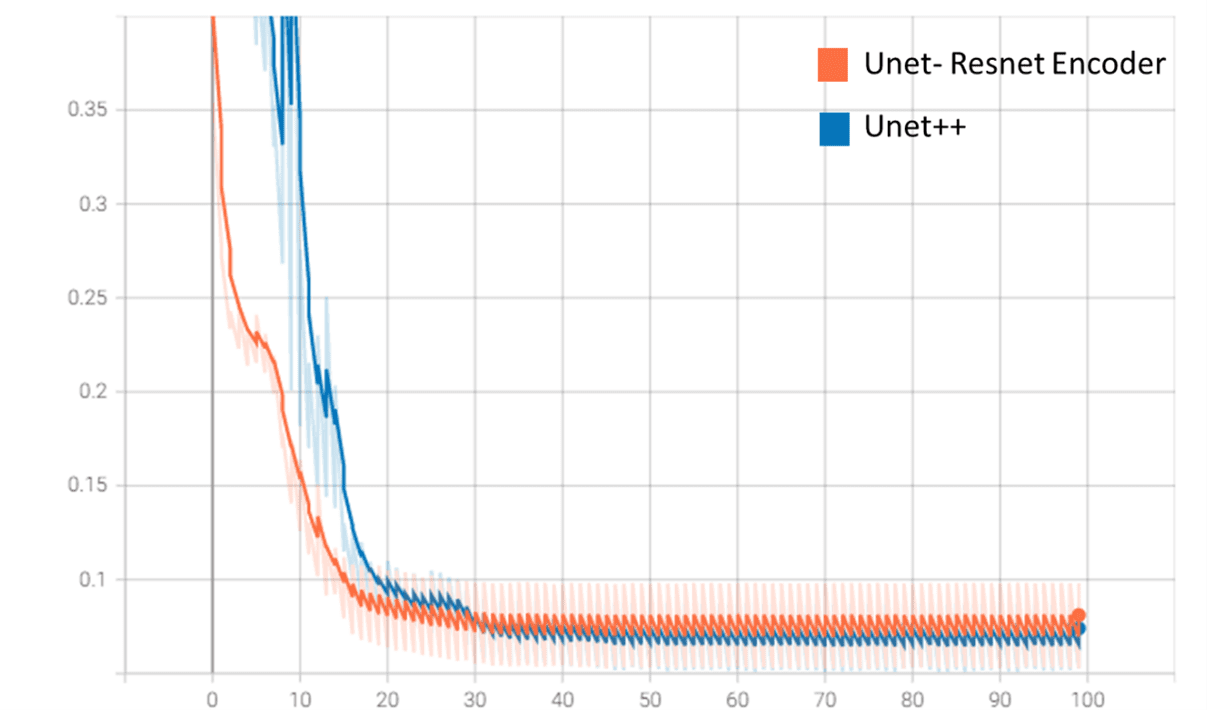
Loss function values (validation) on Tensorboard
On the other hand, although the Unet++ model performs better in validation (loss function “tells”), Unet-Resnet trains much faster and has a smaller size (70 MB vs. 500 MB for Unet++).
Several results of the model using the validation data set are shown below. Each example consists of the input image, the image labeled by the physician/specialist, and the prediction made by the model.
Color coding: Orange – Spleen, Yellow – Right Kidney, Brown – Left Kidney, Blue – Liver.
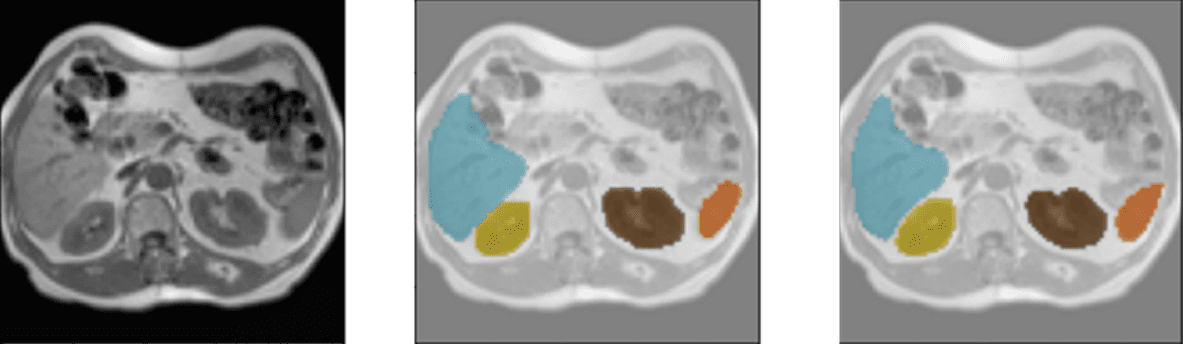
Detection of liver, spleen, and kidneys
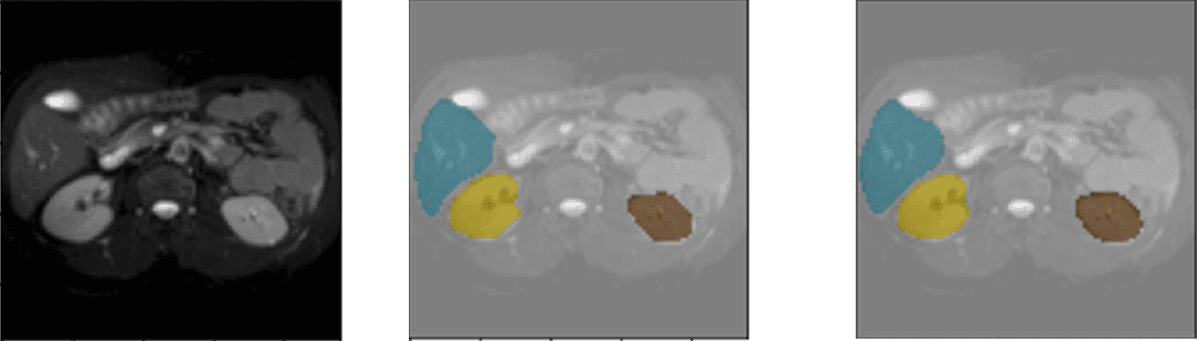
Liver and kidney screening
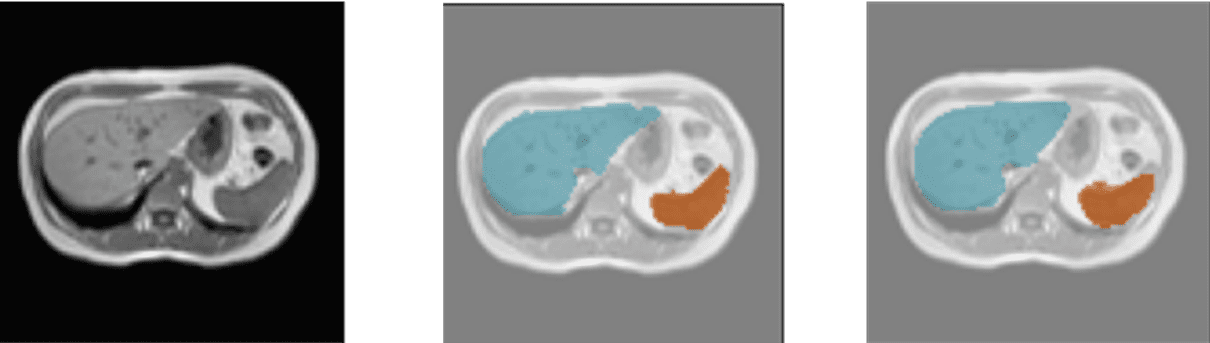
Liver and spleen detection
The use of Deep Learning techniques in medical imaging can help specialists in their decision-making when working on a diagnosis.
One of the first tasks that can facilitate the use of this type of techniques is the integration of the model in any of the hardware devices used by specialists to visualize or represent medical images (mobile devices, HoloLens, and computers). To carry out this process, it will be necessary to previously evaluate the performance of the model and apply different optimization techniques such as: “Pruning or quantization”. The application of these methods will ensure that our model can better adapt to the possible hardware limitations of the device.
As we have seen in this article, our model is able to detect organs from 2D medical images. As a possible future development, this model could be integrated into devices that allow interaction with a 3D representation of the abdomen.
The use of our model within these solutions will help to improve a variety of clinical tests. The system can show at all times where the organs are located and thus, be able to plan the optimal puncture that produces the least amount of damage to adjacent organs.
Elena Canorea
Communications Lead
| Cookie | Duration | Description |
|---|---|---|
| __cfduid | 1 year | The cookie is used by cdn services like CloudFare to identify individual clients behind a shared IP address and apply security settings on a per-client basis. It does not correspond to any user ID in the web application and does not store any personally identifiable information. |
| __cfduid | 29 days 23 hours 59 minutes | The cookie is used by cdn services like CloudFare to identify individual clients behind a shared IP address and apply security settings on a per-client basis. It does not correspond to any user ID in the web application and does not store any personally identifiable information. |
| __cfduid | 1 year | The cookie is used by cdn services like CloudFare to identify individual clients behind a shared IP address and apply security settings on a per-client basis. It does not correspond to any user ID in the web application and does not store any personally identifiable information. |
| __cfduid | 29 days 23 hours 59 minutes | The cookie is used by cdn services like CloudFare to identify individual clients behind a shared IP address and apply security settings on a per-client basis. It does not correspond to any user ID in the web application and does not store any personally identifiable information. |
| _ga | 1 year | This cookie is installed by Google Analytics. The cookie is used to calculate visitor, session, campaign data and keep track of site usage for the site's analytics report. The cookies store information anonymously and assign a randomly generated number to identify unique visitors. |
| _ga | 1 year | This cookie is installed by Google Analytics. The cookie is used to calculate visitor, session, campaign data and keep track of site usage for the site's analytics report. The cookies store information anonymously and assign a randomly generated number to identify unique visitors. |
| _ga | 1 year | This cookie is installed by Google Analytics. The cookie is used to calculate visitor, session, campaign data and keep track of site usage for the site's analytics report. The cookies store information anonymously and assign a randomly generated number to identify unique visitors. |
| _ga | 1 year | This cookie is installed by Google Analytics. The cookie is used to calculate visitor, session, campaign data and keep track of site usage for the site's analytics report. The cookies store information anonymously and assign a randomly generated number to identify unique visitors. |
| _gat_UA-326213-2 | 1 year | No description |
| _gat_UA-326213-2 | 1 year | No description |
| _gat_UA-326213-2 | 1 year | No description |
| _gat_UA-326213-2 | 1 year | No description |
| _gid | 1 year | This cookie is installed by Google Analytics. The cookie is used to store information of how visitors use a website and helps in creating an analytics report of how the wbsite is doing. The data collected including the number visitors, the source where they have come from, and the pages viisted in an anonymous form. |
| _gid | 1 year | This cookie is installed by Google Analytics. The cookie is used to store information of how visitors use a website and helps in creating an analytics report of how the wbsite is doing. The data collected including the number visitors, the source where they have come from, and the pages viisted in an anonymous form. |
| _gid | 1 year | This cookie is installed by Google Analytics. The cookie is used to store information of how visitors use a website and helps in creating an analytics report of how the wbsite is doing. The data collected including the number visitors, the source where they have come from, and the pages viisted in an anonymous form. |
| _gid | 1 year | This cookie is installed by Google Analytics. The cookie is used to store information of how visitors use a website and helps in creating an analytics report of how the wbsite is doing. The data collected including the number visitors, the source where they have come from, and the pages viisted in an anonymous form. |
| attributionCookie | session | No description |
| cookielawinfo-checkbox-analytics | 1 year | Set by the GDPR Cookie Consent plugin, this cookie is used to record the user consent for the cookies in the "Analytics" category . |
| cookielawinfo-checkbox-necessary | 1 year | This cookie is set by GDPR Cookie Consent plugin. The cookies is used to store the user consent for the cookies in the category "Necessary". |
| cookielawinfo-checkbox-necessary | 11 months | This cookie is set by GDPR Cookie Consent plugin. The cookies is used to store the user consent for the cookies in the category "Necessary". |
| cookielawinfo-checkbox-necessary | 11 months | This cookie is set by GDPR Cookie Consent plugin. The cookies is used to store the user consent for the cookies in the category "Necessary". |
| cookielawinfo-checkbox-necessary | 1 year | This cookie is set by GDPR Cookie Consent plugin. The cookies is used to store the user consent for the cookies in the category "Necessary". |
| cookielawinfo-checkbox-non-necessary | 11 months | This cookie is set by GDPR Cookie Consent plugin. The cookies is used to store the user consent for the cookies in the category "Non Necessary". |
| cookielawinfo-checkbox-non-necessary | 11 months | This cookie is set by GDPR Cookie Consent plugin. The cookies is used to store the user consent for the cookies in the category "Non Necessary". |
| cookielawinfo-checkbox-non-necessary | 11 months | This cookie is set by GDPR Cookie Consent plugin. The cookies is used to store the user consent for the cookies in the category "Non Necessary". |
| cookielawinfo-checkbox-non-necessary | 1 year | This cookie is set by GDPR Cookie Consent plugin. The cookies is used to store the user consent for the cookies in the category "Non Necessary". |
| cookielawinfo-checkbox-performance | 1 year | Set by the GDPR Cookie Consent plugin, this cookie is used to store the user consent for cookies in the category "Performance". |
| cppro-ft | 1 year | No description |
| cppro-ft | 7 years 1 months 12 days 23 hours 59 minutes | No description |
| cppro-ft | 7 years 1 months 12 days 23 hours 59 minutes | No description |
| cppro-ft | 1 year | No description |
| cppro-ft-style | 1 year | No description |
| cppro-ft-style | 1 year | No description |
| cppro-ft-style | session | No description |
| cppro-ft-style | session | No description |
| cppro-ft-style-temp | 23 hours 59 minutes | No description |
| cppro-ft-style-temp | 23 hours 59 minutes | No description |
| cppro-ft-style-temp | 23 hours 59 minutes | No description |
| cppro-ft-style-temp | 1 year | No description |
| i18n | 10 years | No description available. |
| IE-jwt | 62 years 6 months 9 days 9 hours | No description |
| IE-LANG_CODE | 62 years 6 months 9 days 9 hours | No description |
| IE-set_country | 62 years 6 months 9 days 9 hours | No description |
| JSESSIONID | session | The JSESSIONID cookie is used by New Relic to store a session identifier so that New Relic can monitor session counts for an application. |
| viewed_cookie_policy | 11 months | The cookie is set by the GDPR Cookie Consent plugin and is used to store whether or not user has consented to the use of cookies. It does not store any personal data. |
| viewed_cookie_policy | 1 year | The cookie is set by the GDPR Cookie Consent plugin and is used to store whether or not user has consented to the use of cookies. It does not store any personal data. |
| viewed_cookie_policy | 1 year | The cookie is set by the GDPR Cookie Consent plugin and is used to store whether or not user has consented to the use of cookies. It does not store any personal data. |
| viewed_cookie_policy | 11 months | The cookie is set by the GDPR Cookie Consent plugin and is used to store whether or not user has consented to the use of cookies. It does not store any personal data. |
| VISITOR_INFO1_LIVE | 5 months 27 days | A cookie set by YouTube to measure bandwidth that determines whether the user gets the new or old player interface. |
| wmc | 9 years 11 months 30 days 11 hours 59 minutes | No description |
| Cookie | Duration | Description |
|---|---|---|
| __cf_bm | 30 minutes | This cookie, set by Cloudflare, is used to support Cloudflare Bot Management. |
| sp_landing | 1 day | The sp_landing is set by Spotify to implement audio content from Spotify on the website and also registers information on user interaction related to the audio content. |
| sp_t | 1 year | The sp_t cookie is set by Spotify to implement audio content from Spotify on the website and also registers information on user interaction related to the audio content. |
| Cookie | Duration | Description |
|---|---|---|
| _hjAbsoluteSessionInProgress | 1 year | No description |
| _hjAbsoluteSessionInProgress | 1 year | No description |
| _hjAbsoluteSessionInProgress | 1 year | No description |
| _hjAbsoluteSessionInProgress | 1 year | No description |
| _hjFirstSeen | 29 minutes | No description |
| _hjFirstSeen | 29 minutes | No description |
| _hjFirstSeen | 29 minutes | No description |
| _hjFirstSeen | 1 year | No description |
| _hjid | 11 months 29 days 23 hours 59 minutes | This cookie is set by Hotjar. This cookie is set when the customer first lands on a page with the Hotjar script. It is used to persist the random user ID, unique to that site on the browser. This ensures that behavior in subsequent visits to the same site will be attributed to the same user ID. |
| _hjid | 11 months 29 days 23 hours 59 minutes | This cookie is set by Hotjar. This cookie is set when the customer first lands on a page with the Hotjar script. It is used to persist the random user ID, unique to that site on the browser. This ensures that behavior in subsequent visits to the same site will be attributed to the same user ID. |
| _hjid | 1 year | This cookie is set by Hotjar. This cookie is set when the customer first lands on a page with the Hotjar script. It is used to persist the random user ID, unique to that site on the browser. This ensures that behavior in subsequent visits to the same site will be attributed to the same user ID. |
| _hjid | 1 year | This cookie is set by Hotjar. This cookie is set when the customer first lands on a page with the Hotjar script. It is used to persist the random user ID, unique to that site on the browser. This ensures that behavior in subsequent visits to the same site will be attributed to the same user ID. |
| _hjIncludedInPageviewSample | 1 year | No description |
| _hjIncludedInPageviewSample | 1 year | No description |
| _hjIncludedInPageviewSample | 1 year | No description |
| _hjIncludedInPageviewSample | 1 year | No description |
| _hjSession_1776154 | session | No description |
| _hjSessionUser_1776154 | session | No description |
| _hjTLDTest | 1 year | No description |
| _hjTLDTest | 1 year | No description |
| _hjTLDTest | session | No description |
| _hjTLDTest | session | No description |
| _lfa_test_cookie_stored | past | No description |
| Cookie | Duration | Description |
|---|---|---|
| loglevel | never | No description available. |
| prism_90878714 | 1 month | No description |
| redirectFacebook | 2 minutes | No description |
| YSC | session | YSC cookie is set by Youtube and is used to track the views of embedded videos on Youtube pages. |
| yt-remote-connected-devices | never | YouTube sets this cookie to store the video preferences of the user using embedded YouTube video. |
| yt-remote-device-id | never | YouTube sets this cookie to store the video preferences of the user using embedded YouTube video. |
| yt.innertube::nextId | never | This cookie, set by YouTube, registers a unique ID to store data on what videos from YouTube the user has seen. |
| yt.innertube::requests | never | This cookie, set by YouTube, registers a unique ID to store data on what videos from YouTube the user has seen. |
